What are LRGs?
A Locus Reference Genomic (LRG) is a manually curated record that contains stable and thus, un-versioned reference sequences designed specifically for reporting sequence variants with clinical implications.
Accurate and unambiguous reporting of variants requires internationally recognized reference sequences that do not change over time. The use of multiple sequences for a given locus as well as confusion over versions has resulted in inconsistent variant reporting in the past. The LRG project was created to avoid these problems.
Each LRG contains a stable “fixed” section and a regularly updated “updatable” section. The fixed section contains stable genomic DNA sequence for the region of interest, transcripts and proteins deemed essential for reporting variants, and an LRG-specific exon numbering system. The updatable section contains mapping information, annotation of all transcripts and overlapping genes in the regions, and other relevant information submitted by the community (e.g legacy exon numbering).
During the LRG creation process, LRG curators will review the transcript submitted by the requester as well as all other transcripts in the region of interest for potential inclusion in the record. Curators will perform alignments, review supporting evidence for each transcript and analyze expression data as part of this review process. Only transcripts for which there is currently good biological understanding AND are required for the unambiguous reporting of disease-causing variants will be included.
The MANE (Matched Annotation from the NCBI and EMBL-EBI) Project is a joint initiative between EMBL-EBI’s Ensembl/GENCODE Project and NCBI’s RefSeq project. MANE aims to release a genome-wide transcript set that contains one well-supported transcript per protein-coding locus (MANE Select). All transcripts in the MANE set perfectly align to GRCh38 and represent 100% identity (5’UTR, coding sequence, 3’UTR) between the RefSeq (NM) and corresponding Ensembl (ENST) transcript.
The flag MANE Select has been included in the records to note which transcript is the MANE Select for each gene. Currently, a MANE Select transcript has been defined for ~67% of protein-coding loci, though we aim to achieve genome-wide coverage.
For more information on MANE see Ensembl’s MANE blog and NCBI’s MANE blog.
LRG and RefSeqGene are collaborative resources. The advantages of using LRGs for variant reporting are:
- LRGs are specifically created for the reporting of clinically relevant variants and hence, are for loci with clinical implications.
- LRGs are stable and therefore are not versioned, thus reducing ambiguity when reporting variants.
When an LRG is established for any gene, the RefSeqGene and its annotation will be “frozen” to match that of the LRG.
The sequences in the LRG record do not necessarily perfectly match the reference genome assemblies (GRCh37/38). LRG sequences are based on RefSeqGene sequences, which, if possible, represent prevalent “standard” alleles at each locus. Therefore, there are a number of cases where the RefSeqGene sequences may differ from the reference genome assembly.
-
If the current reference assembly is not well supported an alternate sequence is selected, in consultation with gene-specific experts as available. When feasible, RefSeqGene sequences will be derived from a single clone, based on the assumption that no sequence errors were introduced in cloning, and that a single insert represents an example of a naturally occurring haplotype.
-
The default implementation of ‘standard allele’ is the sequence from the public reference genome assembly. If, however, there is published evidence, evidence from locus-specific databases, or evidence from clinical testers, that the sequence in the reference genome assembly is not standard, the RefSeqGene sequence can be constructed from an alternate source sequence, or locally modified.
During the curation of an LRG record our curators review all mismatches between the RefSeqGene and the reference assemblies (GRCh37 and GRCh38).
Read more about the RefSeqGene project, and its relationship with the LRG.
LRGs are created for the benefit of the biomedical community and so must meet its needs. We welcome discussion about whether or not individual LRGs fulfil specific needs and we will work with the community to ensure that these needs are met. In the end, we will take the authoritative advice of the community.
Once a LRG record has been created, you can e.g.:
-
View your LRG and all known variants in genome browsers supporting LRGs:
-
Ensembl
Once an LRG has been released, it is integrated into the next Ensembl release, normally within two months. Public LRGs can be viewed in a set of dedicated pages (for example, LRG_1) whereas pending LRGs can be viewed by following the Ensembl link in the updatable section of each LRG (for example, LRG_750). -
NCBI
The records are available from NCBI’s Nucleotide and Gene databases and can be found in the RefSeqGene browser page and FTP site (ftp://ftp.ncbi.nlm.nih.gov/refseq/H_sapiens/RefSeqGene/). More information on ways to find LRGs at NCBI can be found here https://www.ncbi.nlm.nih.gov/refseq/rsg/lrg/. -
UCSC
The records are available in the UCSC Genome Browser by entering the LRG identifier or gene symbol in the search box or by turning on the LRG track under Genes and Gene Predictions.
-
-
Check your variant data using your LRG:
- Mutalyzer checks that variants described using your LRG follow HGVS conventions - Mutalyzer’s “Name Checker”, “Syntax Checker”, and “Name Generator” ensure that variants described using LRG sequences follow HGVS guidelines.
- VariantValidator
-
Interpret your variant data using your LRG:
- Variant Effect Predictor - VEP (Ensembl)
- Alamut (Interactive Biosoftware)
- Variobox
The LRG-specific exon numbering system included in each LRG is based on the transcript(s) included in the fixed section. Each exon is numbered consecutively 5′ to 3′; the numbering is then applied to individual transcripts.
Over LRGs have been created, of which are public. The aim is to create an LRG for every locus with clinical implications.
The LRG web site http://www.lrg-sequence.org/ and the partner RefSeqGene site http://www.ncbi.nlm.nih.gov/RefSeq/RSG/ maintain current information about the LRG project and available sequences.
The LRG standard and why it is needed is described in the following publication:
“Locus Reference Genomic: reference sequences for the reporting of clinically relevant sequence variants”
MacArthur JA et al., Nucleic Acids Res. 2014 Jan (doi: 10.1093/nar/gkt1198).
See also “Locus Reference Genomic sequences: an improved basis for describing human DNA variants” (Dalgleish R et al., Genome Med. 2010, 2:24), and the editorial “Conventional wisdom” in Nature Genetics 2010, 42, p.363.
The LRG project is a collaboration between the NCBI and EMBL-EBI. The creation of an LRG involves several steps.
You can check the progress of pending LRGs here.
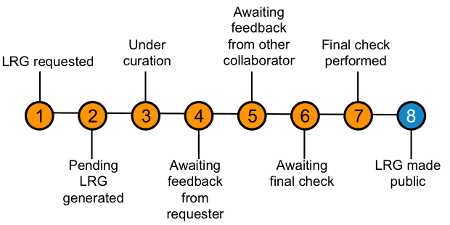
- Request of an LRG Step 1
- LRGs are created upon request by collaborators (e.g. members of diagnostic and research communities and/or clinical experts from specific disease areas, LSDB curators, etc.).
- The genomic sequence included in each LRG record is identical to the RefSeqGene sequence (RSG) for the gene of interest. If there is no RSG for the gene, LRG curators will work with the NCBI to ensure its creation. It is our policy for all LRGs to match the genome reference assembly. However, alternative sequences may be requested if there is a prevalent legacy alternate allele or if there is an error in the genome reference assembly.
- Requesters may suggest any transcript of choice (RefSeq or ENST) for inclusion in an LRG record. If there is no sequence for the desired transcript, LRG curators will work to ensure its creation.
- Creation of pending record Step 2
- Pending records are not finalised and are subject to change.
- Automated Annotation
- Manual Curation Step 3
- Curation of LRG records is an expert activity performed by scientists at the EMBL-EBI.
- Review of genomic reference sequence
- Curators examine the RefSeqGene for any mismatches with respect to the genome reference assembly (GRCh38).
- If a mismatch exists, curators examine the frequency of the alleles
- If the genome reference assembly alleles have a frequency of 5% or less in all populations, LRG curators will report this to GRC and request a review. Genome reference assembly alleles with a frequency higher than 5% in any population are considered representative and will not be requested for review.
- LRG curators also discuss with the requester to determine whether the RefSeqGene alternate alleles represent prevalent legacy alleles.
- The goal is to match the genome reference assembly as much as possible, and curators work with the NCBI to make changes to the RefSeqGene if appropriate.
- Review of Transcripts
- All known transcripts (RefSeq and GENCODE) are manually reviewed by our curators.
- Transcriptional and translational supporting evidence and community usage is assessed.
- LRG curators request that GENCODE and/or RefSeq review the locus of interest if there are significant discrepancies between transcripts.
- Selection of Transcript(s) for inclusion
- We aim to include one transcript, though sometimes more than one will be necessary to ensure that all variants at the locus can be reported.
- Only transcripts with good biological evidence that are essential for reporting variants will be included.
- For each selected transcript, it is our goal to build complete identity between the RefSeq transcript and the corresponding GENCODE transcript.
- Expert review Steps 4 & 5
- Experts in the LRG region of interest are contacted for input. For more on experts see the LRG collaborators page.
- Publication of LRG Steps 6 & 7 Step 8
- Once the LRG record and sequences have been reviewed and approved by requesters, experts and LRG curators, the record is finalised and collaborators informed. These records are referred to as “public” LRGs. The reference sequences included in the fixed section cannot be changed once the LRG is made public.
The updatable section of all records, “pending” and “public”, is refreshed in an automated manner on a weekly basis.
Stability issues
No changes to the sequences in an LRG will be permitted.
If it’s no longer possible to describe a sequence variant in terms of an existing LRG, it might be necessary to create a totally new LRG with a uniquely different number (e.g. LRG_1275 instead of the existing LRG_89). The original LRG will not be “retired” and it will remain valid to describe variants with respect to that sequence record. Creation of additional LRGs for an existing gene or genomic region will only be considered in the most exceptional circumstances.
The RefSeq project, following the convention of the International Nucleotide Sequence Database collaboration (http://www.insdc.org/), assigns sequence identifiers as a combination of a stable component (the accession), and a version. Any revision of the sequence results in the incrementing of the version number. The version number is indicated after the decimal point at end of the accession number (e.g. NM_000088.3).
Unfortunately, the version of a sequence is often not reported when a variant description is presented in a publication. Thus, uncertainty can result when trying to interpret the consequence of any variant if the current version of the reference sequence is greater than 1. This problem is avoided in the LRG accessioning system by not having versions. Once an LRG is created, the sequence data are never changed.
It’s inevitable that new transcripts of biological importance will be discovered for genes for which an LRG already exists. Such transcripts will be included in the updatable section of the record. However, new transcripts can only be added to the fixed section if they are essential for reporting clinically relevant variants. A compelling case can be made for their inclusion if the new transcripts encode different proteins of clear clinical importance and variants cannot be meaningfully described in terms of the current transcripts in the LRG. Consideration of requests for the addition of new transcripts will be on a case-by-case basis.
Versioning is an issue with traditional reference sequence records simply because the actual sequences differ from version to version for records with the same accession number.
The LRG sequence data for the genomic DNA, the transcripts and their translation products will never be changed.
Consequently, a variant description such as LRG_13:g.8290C>A will always remain valid and will never be subject to misinterpretation. The user simply needs to ensure that the LRG contains all of the necessary transcripts for the intended task.
Once a new assembly is released, the mapping information and annotation of all LRGs will be updated to the new assembly. Mapping of the LRG genomic sequence to both the current and penultimate assembly will be included in each LRG.
Once an LRG has been made public, the transcript(s) included in the Fixed section of the record cannot be removed.
This means that if the knowledge about an LRG transcript has changed significantly (i.e. changes to the position of the start ATG or changes to protein coding exons), it is not possible to note this information in the LRG record.
The only way to provide accurate information about an LRG transcript that has changed is to create a new record that includes the updated version of the transcript.
If your favourite LRG has been superseded and you would like more information, please contact us at contact@lrg-sequence.org.
Variant Reporting Standards
No, the standard HGVS Nomenclature will still be used. HGVS and EMQN best practice guidelines have endorsed LRGs.
The stable identifiers of the genomic, transcript, and protein sequences in the fixed section of an LRG (transcripts: “t1”, “t2”, etc.; proteins: “p1”, “p2”, etc.) can be used for stable reporting of variants. See “Can you give me an example?” for more details.
The COL1A1 gene is represented by LRG record LRG_1 which has a single transcript (t1) and a single corresponding protein (p1).
The frequently reported disease-causing variant NG_007400.1:g.9595G>A can also be reported as NM_000088.3:c.769G>A, and as NP_000079.2:p.Gly257Arg using the current RefSeqGene and RefSeq mRNA and protein reference sequences. Since LRGs contain the genomic DNA, mRNA and protein sequences within a single record, the three corresponding descriptions are LRG_1:g.9595G>A, LRG_1t1:c.769G>A and LRG_1p1:p.Gly257Arg.
| Description Level | RefSeqGene or RefSeq | LRG |
|---|---|---|
| Gene | NG_007400.1:g.9595G>A | LRG_1:g.9595G>A |
| mRNA | NM_000088.3:c.769G>A | LRG_1t1:c.769G>A |
| Protein | NP_000079.2:p.Gly257Arg | LRG_1p1:p.Gly257Arg |
The calcitonin gene (CALCA) encodes two peptide hormones, calcitonin and calcitonin gene-related peptide (CGRP), that have no amino acid sequence in common. These hormones are derived by enzymatic cleavage of the translation products of two alternatively spliced mRNAs that exclusively contain exon 4 (calcitonin) or exons 5 and 6 (CGRP). Consequently, a SNP in the first base of exon 4 (rs5241) affects only the mRNA that encodes calcitonin.
Using HGVS nomenclature, the variant can be described as NM_001033952.2:c.228C>A using the calcitonin RefSeq mRNA as the reference sequence. The corresponding protein-level description is NP_001029124.1:p.Ser76Arg.
Alternatively, it can be described with respect to the RefSeqGene genomic DNA sequence as NG_015960.1:g.8290C>A.
The LRG for the CALCA gene (LRG_13) contains information for both the major alternatively spliced forms of the gene’s transcripts. Calcitonin and CGRP are represented by transcripts t2 and t1 respectively. Consequently, the SNP can be described at the DNA level as LRG_13: g.8290C>A or LRG_13t2:c.228C>A.
The corresponding protein-level description is LRG_13p2:p.Ser76Arg.
| Description Level | RefSeqGene or RefSeq | LRG |
|---|---|---|
| Gene | NG_015960.1:g.8290C>A | LRG_13:g.8290C>A |
| mRNA | NM_001033952.2:c.228C>A | LRG_13t2:c.228C>A |
| Protein | NP_001029124.1:p.Ser76Arg | LRG_13p2:p.Ser76Arg |
Yes, several tools can be used to convert variant coordinates from other reference sequences into LRG coordinates.
As the LRG consists of genomic, transcript and protein sequences that are linked, the sequence covering the introns are present too.
Below are examples of using HGVS nomenclature to report a variant in an intron e.g.
rs750106647 is LRG_1t1:c.4005+11T>C and rs778417218 is LRG_1t1:c.4005+5G>A.
Copy number variation (CNV) will certainly be an issue, but LRGs are certainly no less well suited to the task of variant description than existing reference sequence records. Requests will be considered for the creation of an LRG representing a particular allele with respect to CNV and we will work with the requesting party to achieve the best practicable solution to represent the allele.
Obtaining existing LRGs and requesting new ones
To find if an LRG already exists, you may use the search function on this page or search the list of all LRGs found here. You may also search for LRGs in Ensembl and NCBI browsers.
New LRGs are no longer generated and and the LRG website was last updated on 31st March 2021. Ensembl and RefSeq transcripts that are specified by the MANE collaboration are preferred for all genes where available to help standardise clinical reporting. https://tark.ensembl.org/web/mane_project/
Viewing LRGs
You can search by e.g. LRG identifier, HGNC gene name, NCBI and Ensembl accession numbers (gene or transcript), gene synonym or LRG status.
Autocompletion will appear when you will start typing. You can search for NCBI and Ensembl accession numbers without using the version number, e.g. NM_000088 instead of NM_000088.3.
Batch search can be carried out by entering a list of identifiers separated by a semi-colon symbol e.g. LRG_1;LRG_3;NM_007294;LRG_45.
All that you need is a web browser such as Firefox, Chrome, Safari, etc. to view the LRGs that are available at the LRG FTP site.
LRGs can also be viewed in the Ensembl, NCBI and UCSC genome browsers.
Within the browser view of a LRG it is possible to display the individual sequences (genomic DNA, transcripts and their translated protein sequences) in FASTA format. This allows copying and pasting of sequences into other applications that support that format.
From NCBI, try the “graphics” display http://www.ncbi.nlm.nih.gov/nuccore/LRG_1?report=graph.
Yes, each LRG can be downloaded from its own page on the LRG website or by following the instructions described on the Data page. If you want to display the downloaded LRG(s) locally on your web browser with the same layout as the LRG website, you need to download the files:
- lrg2html.xsl
- lrg2html.css
- lrg2html.js
- The img directory
and place these in the same directory as the downloaded LRG file(s).
Without these extra files, your web browser will display the LRG data in XML rather than the nicely formatted version that you see when viewing LRGs from the ftp site.
Software support for LRGs
Yes, some of the information from LRG records are available through different web services from the EMBL-EBI EB-eye RESTful service.
See the LRG page Web services for more detailled information.
Here is the list of external softwares supporting LRGs:
- Mutalyzer’s “Name Checker”, “Syntax Checker”, and “Name Generator” ensure that variants described using LRG sequences follow HGVS guidelines.
- VariantValidator
- Alamut (from Interactive Biosoftware)
- Variant Effect Predictor (from Ensembl)
- Variobox facilitate interpretation of variation data described using LRG coordinates.
In addition,the LOVD DNA variation database system supports LRGs.
Anybody can write an application to handle and manipulate sequence data in the LRGs: the LRG format is open and the record schema is freely available.
We would encourage you to make your software free and open and to let us know about it so that we can provide links to your application.
Technical support for the schema is available at contact@lrg-sequence.org.
Specifications and standards
LRGs are created in extensible markup language (XML) format. Each XML file is highly structured and contains all the information pertaining to a single LRG.
The LRG XML schema was created in RELAX NG schema description language and can be downloaded from http://ftp.ebi.ac.uk/pub/databases/lrgex/LRG.rnc. The schema has a date stamp and any changes would be accompanied by a change in the date.
The current version of the schema is Schema 1.9 and its documentation can be found on the LRG FTP site.
The current version of the technical specification document is available at http://ftp.ebi.ac.uk/pub/databases/lrgex/docs/LRG.pdf.
There is a version number and date stamp in the bottom margin to help in tracking changes to the specification.
For an LRG to be an international standard, it must be accessioned by the collaborating groups. If you would like to request an LRG, please contact contact@lrg-sequence.org.
We would encourage you not to create your own LRG records.
Administrative Issues
The creation of LRGs is the joint responsibility of the European Bioinformatics Institute (EMBL-EBI) and the National Center for Biotechnology Information (NCBI).
The LRG concept was developed as a project within the remit of the GEN2PHEN project and was funded for 5 years under the European Community’s Seventh Framework Programme (FP7).
Although funding for GEN2PHEN ended in June 2013, EMBL-EBI and NCBI are fully committed to maintaining the LRG project.